- Summary
- Transcripts
- Subcellular localization
- Properties
- Mass spectrometry
- Structure prediction
- mPPI network
Details for
Summary
-
Description
Transcripts
-
Description
Subcellular localization
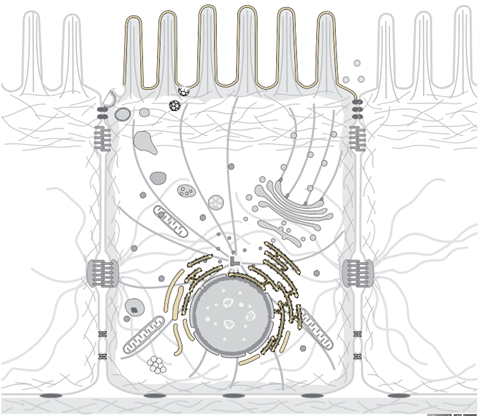
Properties
-
Description
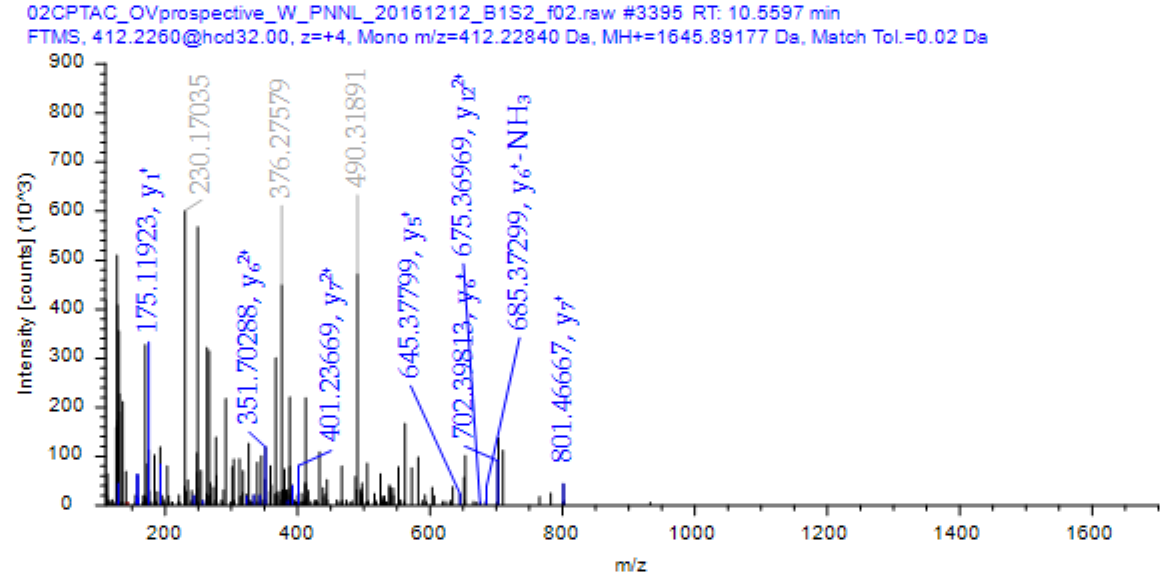
Mass spectrometry
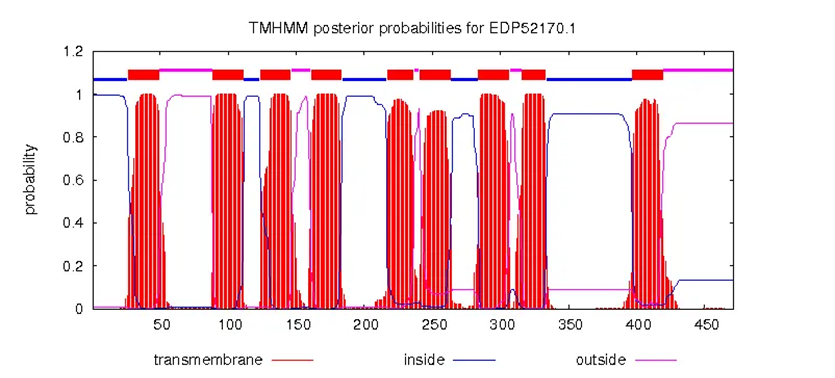
Structure prediction
We used TMHMM-2.0 to predict the possibility of micropeptide transmembrane helices, the results are as follows:
| Length | inside | signal | outside | Number TMRs |
|---|---|---|---|---|
| 45 | / | 1-35 | 36-45 | 0 |
The NetSurfP 3.0 server is used to predict the surface reachability, secondary structure (α-helix, β-strand, random coli), disorder, and phi/psi dihedral Angle of amino acids in amino acid sequences, the results are as follows:
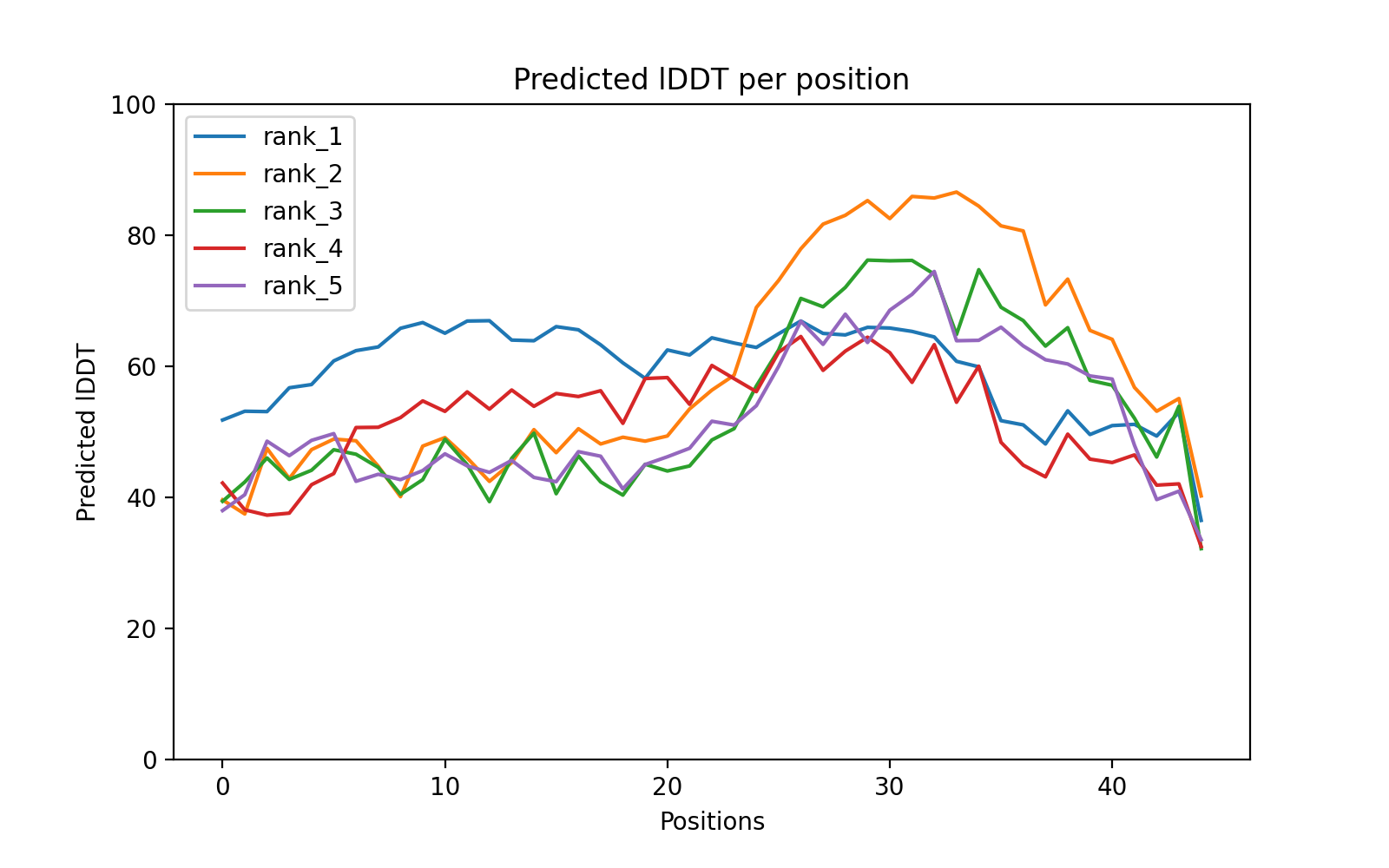
For 3D micropeptide structure prediction, we recommend leveraging sequence-based Alphafold2. Below, we present the structure of “Rank 1” model. For additional details and access to the full set of predictions, kindly refer to the provided download link.

(Click here for downloading all .pdb files)
mPPI network
The micropeptide-protein interaction (mPPI) network represents interactions between micropeptides and established classical proteins. To visually depict the degree of association between micropeptides and proteins, node colors are determined by their scores within the network—nodes with higher scores appear darker, indicating stronger connectivity with other nodes.
Please refer to the diagram below and click to explore these interactions.
Please refer to the diagram below and click to explore these interactions.
(Click here for downloading all .go files)